Introduction:
Mantle Cell Lymphoma (MCL) is a B-cell neoplasm with poor prognosis and high relapse frequency. It follows an heterogenous clinical course and, despite clear improvements in survival among young and fit patients, most patients eventually relapse. Recurrent mutations have been previously described, pinpointing alterations in DNA damage pathways, cell survival and cell-cycle regulation. Nevertheless, the current genomic knowledge of the disease cannot fully explain its high clinical variability. The present study aimed at revealing the mutation profile for the development of personalized treatment approaches in a population-based cohort of patients diagnosed with MCL.
Material & Methods:
We performed sequencing across ~230 lymphomagenesis-related genes on 75 formalin-fixed paraffin-embedded (FFPE) samples from symptomatic patients diagnosed with MCL in Southern Sweden. The panel covered the exonic parts of the chosen genes with capture probes provided by TWIST Bioscience. Sequence alignment was done against the human genome version 37 and variant calling was performed resorting to Sention TNscope. IGV was used for manual validation of each identified mutation. Samples were pre-selected according to DNA quality. Data on patient characteristics, prognostic factors and treatment was extracted from the Swedish Lymphoma Register.
Results:
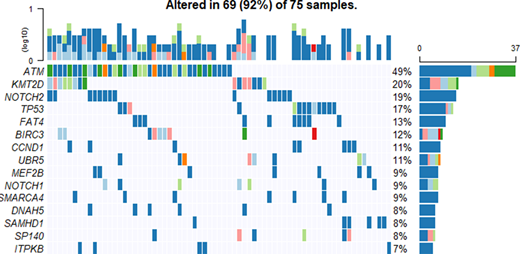
MCL tumors were highly heterogenous with respect to the mutational status of the analyzed genes, ranging between 0 and 45 mutated genes in this cohort. ATM was the most commonly mutated gene (49%), followed by KMT2D, NOTCH2, TP53, FAT4, BIRC3, CCND1, UBR5, MEF2B, NOTCH1,SMARCA4, DNAH5,SAMHD1 and SP140 (Figure 1). Our results are in accordance with the previously published analyses, although the frequency of samples with mutated NOTCH2 was higher (18.7%, compared to 3-12.5% of affected tumors in previous studies), and with less samples with mutated NOTCH3 (6.7% of the studied tumors were mutated, vs 12.5% reported by Yang et al., (2018)). We found mutations in DNAH5, PCLO, FAT3 and BRCA1 genes, which have not been previously reported in MCL. Additionally, FAT4 (14.7%) showed recurrent mutations in our cohort, but has only been referred as commonly mutated in MCL before by Zhang et al. (2014).
Main affected pathways were NOTCH-related and TP53-related pathways, followed by chromatin remodeling and cell cycle pathways. Due to low numbers of patients with non-classic morphology, no mutations were significantly different comparing classical and non-classical histology, although there was an enrichment of TP53 and NOTCH1 mutations in non-classic cases. It seems that cases with non-classic morphology have stronger driver mutations, which may reflect their aggressiveness. Cases with high proliferation (Ki67 >30%) showed a higher frequency of SMARCA4 (30%, vs 7.7% for low proliferation tumors) and TP53 mutations (36% in highly proliferative tumors vs. 17% in low proliferative tumors). No differences in mutation patterns were found in cases with low vs. high SOX11 expression. Unsurprisingly, TP53 mutations were the strongest independent factor for prognosis among the studied genes.
Conclusions:
Using the results from a population-based cohort of MCL, we report the feasibility of applying a targeted panel and sequencing technologies on DNA from formalin fixed tissue, as a method applicable in clinical routine in tailoring treatment approaches for MCL.
Figure 1. Top 15 genes mutated in the cohort studied. Frequency of mutations on each gene is presented on the right panel and in the upper panel the proportion of affected genes by sample can be seen. Darker blue identifies a missense mutation, light blue corresponds to a frameshift deletion, pink color matches frameshift insertions, orange represent in frame deletions, whereas red in frame insertions. Light green identifies nonsense mutations and dark green stands for different types of mutation affecting that gene in that sample.
Jerkeman:Roche: Research Funding; Abbvie: Research Funding; Celgene: Research Funding; Gilead: Research Funding; Janssen: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal